RNA in 3D? Polish scientist leave competition behind!
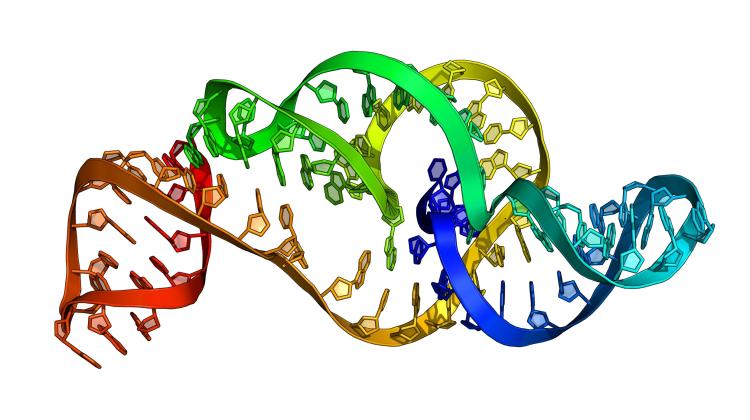 The 3D structure model of the Zika virus RNA generated by RNAComposer on the basis of a sequence. Currently, this structure is already determined experimentally and deposited in the PDB database. Source: Marta Szachniuk
The 3D structure model of the Zika virus RNA generated by RNAComposer on the basis of a sequence. Currently, this structure is already determined experimentally and deposited in the PDB database. Source: Marta Szachniuk
Already 1 million times researchers and people from around the world have used RNAComposer - a publicly available, effective 3D RNA structure modelling system developed in Poznań. And this is not the only Polish success in research on determining the RNA structure.
RNAs are ribonucleic acid molecules. Without them, cells would not be able to produce proteins - molecules that are important for cell structure and function. There are a lot of RNA types and they have different functions in the cell.
For example, matrix RNAs are intermediaries that allow information - a recipe for proteins- to be extracted from the DNA. Ribosome RNAs are built of ribosomes - cellular protein production centres. And transfer RNAs are supposed to bring the appropriate amino acids - building blocks of proteins - to these centres.
RNA functions are so important that scientists from around the world are investigating the properties of its various molecules. However, in order to more effectively predict the functions of a given RNA and to be able to design fully functional molecules in laboratories, one needs to know the 3D structure of a particular RNA, or how it is folded in space.
"The functions of RNA depend on its spatial structure" - said Dr. Marta Szachniuk from the European Centre for Bioinformatics and Genomics (a consortium of the Poznań University of Technology and the Institute of Bioorganic Chemistry PAS).
Meanwhile, however, it is not easy to investigate with experimental methods how individual molecules are folded.
RNA molecule is usually formed by a strand consisting of nucleotide residues joined together (abbreviated: A, C, G, U). Even if their order in the RNA chain is deciphered, i.e. their sequence determined, it is uncertain how the whole molecule is arranged in space. Because RNA molecules - unlike a headphone cable thrown into a backpack - do not tangle into random knots. There are certain rules that allow to predict the shape that the molecule will take. Computer methods for predicting 3D RNA structures are helpful in solving this problem.
Dr hab. Marta Szachniuk together with a team of Prof. Ryszard Adamiak from the Institute of Bioorganic Chemistry PAS in Poznań has developed a free, publicly available system called RNAComposer. An RNA sequence (or information on interactions between nucleotide residues, the so-called secondary structure) is introduced into the system, and in a few/several seconds it calculates and presents a three-dimensional model of the molecule. The program efficiently handles both short and very long chains of RNA molecules with complex architecture.
"Many scientists all over the world use RNAComposer to get the first idea of what their target molecule may look like in 3D. Our system has already made 1 million predictions since 2012" - said Dr. Marta Szachniuk.
This is not the only computer system for predicting 3D RNA structures. There are several such automatic systems. In addition, Besides, research teams conduct experimental research to predict RNA structures.
To compare the effectiveness of different methods of determining the shape of RNA in 3D space, RNA-Puzzles competition has been organized since 2010. The idea is to determine the exact structure of the molecule as accurately as possible with the given RNA sequence. The models provided by the competition participants are then compared with the results of chemical and biological experiments leading to the determination of the structure. The competition is currently organized in two categories: servers that automatically generate results, and human predictions, where models are created as a result of integration of computer calculations and laboratory experiments. "We are the best in the category of automatic 3D RNA prediction systems" - emphasised Dr. Szachniuk.
RNAComposer was created thanks to the fact that for a decade the team from the European Centre for Bioinformatics and Genomics has been building a huge database of RNA data. The data from a huge number of experiments have been collected in the RNA database - FRABASE. Those data allow to draw conclusions about the spatial structures of RNA molecules. This database is still being updated and everyone can use it free of charge. "It is a popular tool. We even know that, for example, at foreign universities students use it as part of research and studies preparing for the profession of bioinformatics experts or biologists" - said Dr. Szachniuk. This database helps them to search, for example for specific spatial patterns that are repeated in different molecules.
Poland is also visible on the global RNA structure research map because of the research conducted by other teams. An important figure is Prof. Ryszard Kierzek from the Institute of Bioorganic Chemistry PAS in Poznań. His work allowed to determine the thermodynamic rules of RNA folding. Innovative research into the determination of RNA structure is also carried out by the team of Prof. Janusz Bujnicki from the International Institute of Molecular and Cell Biology in Warsaw.
Author: Ludwika Tomala
PAP - Science in Poland
lt/ ekr/ kap/
tr. RL
Przed dodaniem komentarza prosimy o zapoznanie z Regulaminem forum serwisu Nauka w Polsce.















